BACKGROUNDHigh-throughput genotyping platforms play vital roles in plant genomic research. Cotton (Gossypium spp.) is the world’s vital pure textile fiber and oil crop.
Upland cotton accounts for greater than 90% of the world’s cotton manufacturing, nonetheless, trendy upland cotton cultivars have slim genetic variety.
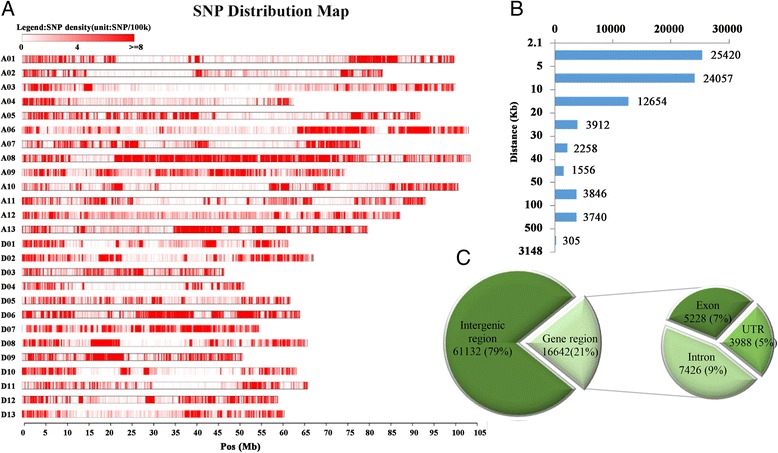
The quantities of genomic sequencing and re-sequencing knowledge launched make it potential to develop a high-quality single nucleotide polymorphism (SNP) array for intraspecific genotyping detection in cotton.
RESULTSHere we report a high-throughput CottonSNP80K array and its utilization in genotyping detection in numerous cotton accessions.
82,259 SNP markers have been chosen from the re-sequencing knowledge of 100 cotton cultivars and used to supply the array on the Illumina Infinium platform. 77,774 SNP loci (94.55%) have been efficiently synthesized on the array. Of them, 77,252 (99.33%) had name charges of>>95% in 352 cotton accessions and 59,502 (76.51%) have been polymorphic loci.
Application exams utilizing 22 cotton accessions with dad or mum/F1 combos or with related genetic backgrounds confirmed that CottonSNP80K array had excessive genotyping accuracy, good repeatability, and large applicability.
Phylogenetic evaluation of 312 cotton cultivars and landraces with large geographical distribution confirmed that they might be labeled into ten teams, irrelevant of their origins. We discovered that the totally different landraces have been clustered in numerous subgroups, indicating that these landraces have been main contributors to the event of various breeding populations of contemporary G. hirsutum cultivars in China.
We built-in a whole of 54,588 SNPs (MAFs>>0.05) related to 10 salt stress traits into 288 G. hirsutum accessions for genome-wide affiliation research (GWAS), and eight important SNPs related to three salt stress traits have been detected.
CONCLUSIONSWe developed CottonSNP80K array with excessive polymorphism to differentiate upland cotton accessions. Diverse utility exams indicated that the CottonSNP80K play vital roles in germplasm genotyping, selection verification, practical genomics research, and molecular breeding in cotton.

Genomic analyses of tropical beef cattle fertility based mostly on genotyping swimming pools of Brahman cows with unknown pedigree.
We introduce an revolutionary strategy to decreasing the general price of acquiring genomic EBV (GEBV) and encourage their use in business intensive herds of Brahman beef cattle.
In our strategy, the DNA genotyping of cow herds from 2 unbiased properties was carried out utilizing a high-density bovine SNP chip on DNA from pooled blood samples, grouped in response to the results of a being pregnant check following their first and second becoming a member of alternatives.
For the DNA pooling technique, 15 to 28 blood samples from the identical phenotype and up to date group have been allotted to swimming pools.
Across the two properties, a whole of 183 swimming pools have been created representing 4,164 cows. In addition, blood samples from 309 bulls from the identical properties have been additionally taken. After genotyping and high quality management, 74,584 remaining SNP have been used for analyses. Pools and particular person DNA samples have been associated by the use of a “hybrid” genomic relationship matrix.
The pooled genotyping evaluation of two massive and unbiased business populations of tropical beef cattle was in a position to get well important and believable associations between SNP and being pregnant check end result. We focus on 24 SNP with important affiliation ( < 1.0 × 10) and mapped inside 40 kb of an annotated gene.
We have established a technique to estimate the GEBV in younger herd bulls for a trait that is at present unable to be predicted in any respect. In abstract, our novel strategy allowed us to conduct genomic analyses of fertility in 2 massive business Brahman herds managed underneath intensive pastoral circumstances.
